Clonal evolution and the ‘survival of the nastiest’ remain the chief obstacles to curing cancer. But what if we could find a way to use the principles of evolution to beat evolving cancers cells at their own game? Simon Crompton explores cutting edge efforts to do just that.
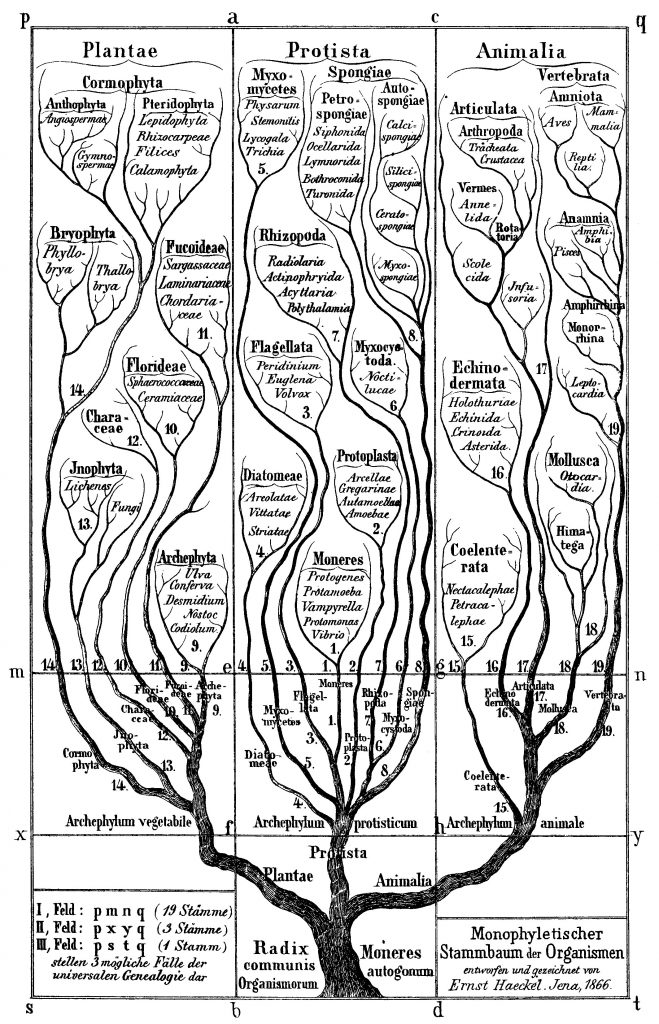
As notebook scrawls go, this one was earth-shaking. In 1837, 12 years before his book On the Origin of Species was published, Charles Darwin sketched a spidery tree depicting how evolution might work, and wrote the words “I think” above it. This was the beginning of what became known as Darwin’s tree of life – and it forms the basis for our understanding of species evolution to this day.
What Darwin might not have predicted in 1837 was that, here in the 21st century, his tree of life would also be forming the basis of a new understanding of the way cancers advance. Today, a group of innovative scientists are using Darwinian principles to not only understand the daunting genetic complexity of advancing and metastasised tumours, but also to devise innovative approaches to controlling them.
The new wave of interest in Darwinian principles has been spearheaded by Charles Swanton, Chair in Personalised Cancer Medicine at University College London’s Cancer Institute, and leader of the research group at the Francis Crick Institute examining genetic diversity in cancer.
His research has indicated not only that single biopsy samples are likely to severely underestimate the genetic variety of cells within tumours, but also that this heterogeneity will nearly always lead to the failure of therapies that target specific types of cell.
His first paper demonstrating the extent of heterogeneity, published in the New England Journal of Medicine in 2012, has been cited more than 3,000 times in the past four years, and prompted an unprecedented number of publications focusing on the evolutionary processes that cause such a diverse ‘fauna’ of cancer cells within a single tumour.
On the one hand, this new understanding of the branched evolutionary progress of advanced cancers provides a bleak analysis of why so many treatments ultimately fail. But on the other, it gives researchers and clinicians a new and firm grounding on which to constructively face their continual frustrations about treatment resistance, setting a new agenda for investigating new and potentially effective treatment options. Researchers have now embarked on work finding ways to harness evolutionary forces to control competing cells, or to cut off advanced cancers at their evolutionary trunk.
“I’m massively optimistic about the prospects, but we’re engaged in a battle of wits with evolution,” says Swanton, a practising oncologist at University College London Hospital, as well as one of the UK’s leading cancer researchers. In November 2016 he won the Biochemical Society’s GlaxoSmithKline Award in recognition of research leading to new advances in medical science.
Cancer Research UK is supporting the work of Swanton and his team, and has invested £14 million into an ambitious national collaboration between six clinical centres and four science centres, to track and understand the evolutionary genetic changes in non-small-cell lung cancer over time in 850 patients (the TRACERx study).
But it isn’t just Cancer Research UK that is convinced of the importance of understanding cancer evolution. The Institute of Cancer Research has just established a new Centre for Evolution and Cancer, led by Mel Greaves. “We have the objective of applying evolutionary principles to forge what we think is a paradigm shift in how we think about and understand cancer,” says Greaves, who specialises in examining the genetic influences and biological pathways that lead to childhood leukaemia. “The implications for cancer treatment are extraordinary.”
The theory of cancer evolution
Researchers have long known that mutations accumulate as cancers develop. But traditional ways of explaining this process never made sense to Charles Swanton. When he was a medical student 20 years ago, he was taught that cancers evolve in a linear manner.
The theory went that a normal cell acquires a mutation – say to the APC gene – that allows that cell to proliferate, dominate other cells and form a tumour. Then one of the cells in the tumour mass also develops a mutation in the p53 gene, and that in turn becomes dominant. Then one of those cells loses chromosome 18, and those cells take over. The process continues, and the tumour grows into a roughly homogenous mass, each cell having the same gene mutations. If that theory were true, wherever you took your biopsy in the tumour, the results of genetic sequencing would be more or less the same.
But when Swanton became a clinician, he couldn’t square this theory with what he saw happening in patients. Why were they becoming resistant to drugs that were targeting the same mutations found in biopsies? He could only think that there must be greater genetic diversity in the tumour than accounted for by linear evolution – that there must nearly always be some cells in the tumour resistant to treatment which would survive and take over.
So his team asked what happened if you performed genetic sequencing on ten biopsies from different parts of a tumour, rather than the customary single biopsy.
“We wanted to know how accurate a picture one biopsy gave you of the tumour genome,” says Swanton. “And the answer is, depending on the type of tumour you’re looking at, not very.” The results were published in his influential 2012 paper in the New England Journal of Medicine, which revealed that in multiple kidney cancer biopsies from the same person, no two samples were the same. For each person studied, Swanton’s team found more than 100 mutations in each tumour sample sequenced, but only one third of them occurred in all samples.
“What’s happening is there’s not linear evolution at all – you very rarely see that. What you see instead is branched evolutionary trajectories of tumours, as Darwin would have predicted, creating tremendous diver-sity from one region of the tumour to another and between primary and metastatic sites.
“So yes, it all comes back to a common ancestor, a single cell back in the history of the tumour, but what’s happened over perhaps ten years is constantly branching evolution has created huge amounts of diversity and robustness, and that’s allowed one or more cells to be resistant to therapy over time.”
That means Darwin’s tree of life can be applied almost exactly to cancers. As Darwin wrote: “The affinities of all the beings of the same class have sometimes been represented by a great tree… As buds give rise by growth to fresh buds, and these, if vigorous, branch out and overtop on all sides many a feebler branch, so by generation I believe it has been with the great Tree of Life, which fills with its dead and broken branches the crust of the earth, and covers the surface with its ever branching and beautiful ramifications.”
Ironically, it is the ever branching and beautiful ramifications of the evolving tree that causes advancing cancer to become untreatable and lethal.
The implications
On the face of it, the implications are depressing. If each tumour has the variety and individuality of a snowflake, are all therapies doomed to fail eventually?
The obvious way of meeting the challenge of resistance is to use combination therapies – targeting two or more mutations at once to try and control disease for much longer periods. There is some evidence that this works in some patients. A modelling exercise led by Bert Vogelstein from Johns Hopkins Kimmel Cancer Center, a pioneer of research into the genetic changes that drive cancer, predicted that dual targeted therapy could result in long-term disease control for most pancreatic, colorectal, and melanoma cancer patients with metastatic disease.
But Swanton believes that turning to combination therapies is impractical for two reasons. First, because every tumour has a unique combination of driving genetic events, finding the right combination of available therapies – and designing the trials to demonstrate effectiveness – would be unfeasibly complicated. Second, all targeted therapies have associated toxicity, and combinations will always be limited by what a patient’s healthy tissue will be able to stand. So Darwinian understanding of tumours provides little prospect of advanced cancer being cured by drugs targeting single mutations.
But it does present hope elsewhere – and a new perspective on how to tackle the infuriating complexity and resilience of cancer.
According to Mel Greaves, Director of the Centre for Cancer and Evolution at the Institute of Cancer Research, there are several areas where evolutionary understanding of cancers offers enormous potential.
“First, the more we understand cancer evolutionary biology, the more we understand how important it is to intervene early: once cancer evolution is up and running there’s a point of no return. Second, it has implications for personalised treatment and targeted medicine: we need to ask whether a target molecule is in every cancer cell or a side branch of an evolutionary clone. Ideally we should be targeting mutations in the trunk of the tree.
“This heterogeneity will nearly always lead to the failure
of therapies that target specific types of cell”
“The third point is whether we can envisage a Darwinian by-pass – directing our approach not directly at cancer cells but towards their micro-environmental habitats and changing their habitat and dependencies. Anti-angiogenesis is a prime example of this tactic.
“A further alternative is to seek to control cancer rather than eradicate it, confronting drug resistance in some cells by allowing competitor cells to survive and consume resources that would otherwise benefit resistant clones.”
New approaches: targeting the evolutionary trunk
Given the diversity of cells within a tumour, the overwhelming challenge is to get a treatment that affects all the cancer cells – not just those that have sprung from an evolutionary branch. Targeting the mutations where it all started and which are present in every cell – the trunk of the evolutionary tree – is the obvious way to fell the entire structure.
But this is not as easy as it sounds. Although we know that there are some key driver gene mutations for many cancers, and that some mutations – for example p53 and KRAS – are found in a large proportion of tumours, they have proved very hard to target with small molecules.
“But even if we do find ways of targeting these molecules, I still fear that resistance is inevitable,” says Swanton. “I think we’re going to have much more success exploiting the immune system – the very system which has evolved over four billion years to target the kind of ever-changing diversity that tumours display.”
The reason for Swanton’s optimism about immunotherapy largely lies in the findings of another groundbreaking study carried out by his team at University College London, and published in Science last year. It discovered that all cancer cells have distinctive ‘flags’ on their surface, deriving from multiple trunk mutations. These can help direct the body’s immune system to attack all cancer cells, not just the branch clones.
“Truncal tumour neo-antigens could allow scientists
to target and destroy tumours
without harming healthy tissues”
Immunotherapies help the patient’s disease-fighting T-cells hunt and destroy cancer cells. But despite their immense potential, trials show they work only in a proportion of patients, and they sometimes also damage healthy tissue, causing severe side effects.
The challenge seems to be precision: how do you help the immune system identify and then lock onto the best targets – the cells that are all cancer cells, and that make up most of the tumour? T-cells find their target by locking onto distinctive proteins on the surface of cells (antigens) – so one solution would be to help them find a protein that is on the surface of all cancer cells, a protein that has been passed down the generations of cancer cells from the very first mutated cell at the bottom of the evolutionary trunk.
Analysing data from over 200 patients with two types of lung cancer, Swanton and his team discovered that in every cancer patient there are unique ‘flag’ proteins present on the surface of every cancer cell, and only on cancer cells, which can be used to alert the immune system to attack (Science 2016, 351:1463–69). They are called truncal tumour neo-antigens and they could allow scientists to target and destroy tumours without harming healthy tissue.
Their continuing research is examining why the ‘flags’ are being hidden or protected from the immune system, and how to harness the immune cells that do recognise the targets.
A new treatment route looks possible: identifying truncal tumour neo-antigens from biopsies, then finding and harvesting T-cells within the tumour which recognise these, replicating them in the lab and then injecting them into the patient. “This takes personalised medicine to its absolute limit, where each patient would have a unique, bespoke treatment,” says Swanton.
Such advances might be a way off, and will inevitably be expensive – at least in the short-term. But Darwinian understanding of cancer opens up other avenues too.
New approaches: adaptive therapy
What if researchers took a completely new approach to controlling advanced cancers – not fighting against the branching evolution that drives the cancer, but working with it for the benefit of the patient?
This is exactly the approach that researchers in Florida are taking, in work examining whether low doses of chemotherapy might keep cancer at bay more effectively than trying to destroy the tumour completely with high doses.
The work, led by Robert Gatenby from the H. Lee Moffitt Cancer Center, centres on the evolutionary principle of survival of the fittest. If high dose chemotherapy kills off all the cancer cells that respond to chemotherapy, only those that are resistant to chemotherapy will remain. And, freed of the competition from non-resistant cells, they become fit and free to grow and roam – bringing back the cancer with a vengeance.
Gatenby’s team studied this dynamic in mice being treated with Taxol for two different types of breast cancer. When given standard doses, their tumours initially shrank, but grew back as soon as the treatment stopped. But when the researchers gave an initial high dose followed by progressively lower doses as the tumour responded, the mice lived much longer. Between 60 and 80% of the mice could be weaned off the drug completely over an extended period without suffering relapses.
The research, published in Science Translational Medicine in February 2016, indicates that keeping resistant and non-resistant cells in a delicate balance of competition might be the best way to hold both back – not curing the cancer, but controlling it for long periods. The technique is called adaptive therapy.
“We doctors need to learn from environmental ecology
and cancer evolutionary biologists”
“The evolutionary principles that govern adaptive therapy may be applicable to a wide range of breast cancer treatments including hormonal manipulation and immunotherapy, although they will need to undergo further testing in those settings,” says Robert Gatenby, who is leader of the Cancer Biology and Evolution Programme at Moffitt.
Based on these promising preclinical results, the Moffitt researchers have begun the first clinical trial assessing an adaptive treatment strategy for relapsed prostate cancer patients. It will examine whether the conventional approach of giving the hormone therapy abiraterone at the maximum tolerated dose extends progression-free survival more or less than an adaptive approach. This has particular relevance to African-American men, who tend to develop resistance to hormone therapy more rapidly than other ethnic groups.
The Moffitt scientists aim to use the molecular and clinical data from the trial to develop computer models that might guide adaptive therapy in the future.
New ways of thinking are required
If adaptive therapy based on Darwinian understanding of cancers holds much promise, it will also demand a significant rethink of the way cancer treatments are researched. The expectations of doctors and patients, and the very structure of clinical trials, will have to change, according to Charles Swanton.
The problem is that response rate is currently the key marker of a drug’s efficacy. But with adaptive therapy, the aim is not a spectacular response but keeping the tumour stable. “That’s not going to sit comfortably with clinicians and patients,” says Swanton.
“Traditionally, we want to shrink the tumour as much as possible until you can hardly see it on the scan. Naturally one thinks the less of a tumour is there the better, but maybe that’s not the case. Maybe we need to utilise the drug-sensitive tumour clones to out-compete the drug resistant tumour clones that we have no way of treating.”
If researchers and ultimately clinicians are genuinely going to tap in to the insights that Darwinian theory brings to confronting cancer, they are going to have to learn to think more creatively and more strategically.
“We doctors need to fight evolution,” says Swanton. “We need to think about how we can manage evolution in a very clever way, and most importantly how we can learn from environmental ecology and cancer evolutionary biologists like Robert Gatenby.” The battle of wits with evolution is likely to be a long one, but at least the enemy now stands clear in view.


