Early detection, disease prognosis, a guide to treatment, a key to unlock the secrets of how cancers evolve. Researchers have high hopes for what they can learn from the biological detritus shed by primary tumours and metastases.
 With the explosion in knowledge about how genetic abnormalities in cancer change over time, and the corresponding rise in drugs that target them, comes a big challenge. How can these abnormalities be monitored during the course of the disease, so that the right treatments can be started (and stopped) at the right time? And could abnormalities be detected even before there are symptoms of primary disease, such as in pancreatic cancer, where most patients present with advanced tumours?
With the explosion in knowledge about how genetic abnormalities in cancer change over time, and the corresponding rise in drugs that target them, comes a big challenge. How can these abnormalities be monitored during the course of the disease, so that the right treatments can be started (and stopped) at the right time? And could abnormalities be detected even before there are symptoms of primary disease, such as in pancreatic cancer, where most patients present with advanced tumours?
This is the realm of biomarkers and biopsies, and the good news is that clinicians – traditionally much more cautious than lab scientists – are talking about new non-invasive ‘liquid biopsy’ techniques that could be widely available in as little as two years’ time.
Liquid biopsies are performed on body fluid samples – blood being of most interest but also urine, spinal fluid and others – to look for a wide range of circulating cancer biomarkers, from fragments of DNA and RNA, to ‘vesicles’ containing tumour material, to whole cancer cells.
Circulating DNA is of particular interest now. Unlike the few other biomarkers in use in cancer, such as prostate specific antigen (PSA), circulating tumour DNA has come directly from a particular cancer – it cannot come from anywhere else – and technology has advanced so that it can be analysed at low cost and with good accuracy and more easily than tumour cells. Analysing DNA in fluids is therefore deserving of the ‘biopsy’ term.
In Europe, the first application is now approved by the European Medicines Agency (EMA) and on the market, and provides a good example of one of the main benefits of liquid biopsy. It is for the detection of EGFR-activating mutations in non-small-cell lung cancer, which is an indication for treatment with gefitinib (Iressa), which inhibits these mutations.
Whereas previously patients would not be eligible for gefitinib without evidence of EGFR mutation using conventional biopsy, following a trial, the EMA has extended the label of gefitinib so it can now be used if the mutation has been detected in circulating DNA in blood, and it has approved a test kit, made by Qiagen, to do this.
The advantage is straightforward – in advanced or metastatic lung cancer, for which gefitinib is approved, it can be hard to obtain enough or any tissue for testing from some patients, and even when possible the procedure is invasive and not suitable to use for regular monitoring. The liquid biopsy offers a good alternative, although the study that led to the approval showed that tumour biopsy does give more accurate results.
Gefitinib can now be used after the EGFR mutation has been detected in circulating DNA in blood
Coming of age
Fortunato Ciardiello, professor of medical oncology at Naples University, and president-elect of the European Society of Medical Oncology (ESMO), says EMA approval in the case of the EGFR liquid biopsy indicates the approach is coming of age. “There is a lot of evidence now that, especially in large-burden tumours, circulating DNA and other materials can be used as biomarkers, for two main purposes,” he notes. “First, after removal of the primary tumour, the presence of circulating tumour DNA or cells could be evidence that cancer is still in the body at micro-metastatic sites. So there could be prognostic value in identifying patients at risk of relapse, although we have achieved complete local control of the cancer.
“Second, and of more interest in the research community in the past few years, is using a liquid biopsy as a way to monitor treatment or follow the evolution of the disease. We know that most cancers are heterogeneous, and that sub-clones and different molecular alterations occur, and that conventional biopsy can miss these as they can be in certain parts of the tissue only.
“Blood biopsy can give us a better picture of the abnormalities from both primary and metastatic sites, and if we identify certain changes such as mutations or gene amplifications, this could be a marker of sensitivity or resistance to therapies, and can help us in ‘treat or not-treat’ decision-making.”
While the lung cancer EGFR test is for a snapshot of the presence of an activating mutation for cancer cell growth, Ciardiello says studies in a number of cancers, such as metastatic colorectal and lung, are now taking blood samples over time to detect changes that can indicate resistance to the initial therapy before other signs of relapse or tumour progression emerge.
“We have to be cautious, as most of the work is experimental, although the evidence is good,” he says. “We need standardised methods for exporting DNA and assessing the presence of mutated genes and other alterations. We are moving fast, but liquid biopsies cannot yet substitute for finding abnormalities in tissue. However, with the lung EGFR approval we do have a complementary method, with much more to come.”
Howard ‘Jack’ West, a medical oncologist and director of medical therapeutics in the thoracic oncology programme at the Swedish Cancer Institute in Seattle, US, agrees that liquid biopsies are of particular interest for lung cancer due to challenges of tissue collection, and that addressing molecular variability is now on the agenda. “The more we study and acknowledge tumour heterogeneity, the greater problem we recognise it to be. Before there was an alternative, there was little point in obsessing over this challenge. Even now, serum-based testing isn’t necessarily better, but the sensitivity issues may arguably be counterbalanced by it overcoming tumour heterogeneity.
“Sensitivity issues may arguably be counterbalanced by it overcoming tumour heterogeneity”
“It may well be that as the technology improves – as it has been and likely will continue to – the sensitivity issues will become less, and serum testing may eventually prove superior to tissue collection even if access to tissue weren’t a limiting factor. But this is speculation at this stage.”
Nitzan Rosenfeld, who runs a molecular diagnostics group at Cancer Research UK’s Cambridge Group, explains why DNA fragments, which are thought to come from dead cancer cells, are of such interest. “Biomarker research is challenging as you can be unsure whether what you are measuring comes from the cancer or from other parts of the body. If it’s not completely specific to the cancer, the initial specificity gets diluted by confounding effects from other non-cancer cells,” he says. Proteins and RNA, even if mostly specific to the cancer, can often originate from other cells; a marker such as PSA comes from the prostate but not necessarily prostate cancer, which is a reason it is such a controversial test.
“But DNA stands out, as the cancer specific mutations come only from tumour cells, as far as we know, which makes them exquisitely specific – if you know a tumour has a particular mutation, when you find DNA in the blood that has that mutation, even at a distance from the tumour, you know it comes from that cancer.”
There is though a lot of other DNA in the blood from non-cancer cells. Rosenfeld notes that 2 ml of plasma may contain more than 10,000 copies of DNA from healthy cells, but in some cases only a few dozen copies of a tumour’s genome. He adds, however, that obtaining and sequencing circulating tumour DNA to significant sensitivity and specificity has been a great success of new technology in the past few years, which may make it more practical as a source of information about the cancer than circulating whole cancer cells.
DNA vs whole cancer cells
In blood of people with advanced breast cancer there are around 100 times more copies of the cancer genome present as fragments of DNA than there are intact cancer cells, says Rosenfeld, adding that, “It’s harder to analyse cells. They are much more fragile than DNA, which is a robust molecule.” Cells first require mechanical methods for isolating them and special detection techniques, and there is currently only one system approved for use in the US (CellSearch).
Circulating tumour cells are, however, also a strong research area, and probably more studied than DNA so far, with new analysis technology being developed. In breast cancer, a certain threshold of circulating cells has been shown to be prognostic for worse outcomes. There is also research similar to that using DNA fragments that is looking at how information from circulating cells can be used to monitor the impact of treatments and the genetic evolution of tumours over time.
Indeed, there is some debate between protagonists of DNA and cells about which is best. West adjudicates: “It remains to be seen whether circulating DNA or cells will emerge as the right platform. So far, more of the recent trials have shown utility of DNA, and what will determine the winner is what actually works – the proof will be in which delivers the sensitivity in larger trials on a reliable basis, as well as turnaround time, which should ideally be under two weeks.”
“It remains to be seen whether circulating DNA or cells will emerge as the right platform”
Much impetus for the circulating DNA work has come from Bert Vogelstein’s group at Johns Hopkins in the US, Rosenfeld notes. Although knowledge about DNA in the blood dates back to the late 1940s (and circulating tumour cells to the 19th century), other fields such as blood testing for foetal disorders and managing viral infections such as HIV have paved the way for DNA applications in the clinic – cancer has been a late-comer.
In 2008, the team at Johns Hopkins published ‘Circulating mutant DNA to assess tumour dynamics’, which quantified DNA in a small number of patients and showed how a PCR technique pioneered by the group, known as BEAMing, could sequence such DNA.
A lot of companies are now rushing to produce test kits that raise the bar on accuracy (including Inivata, for which Rosenfeld is chief scientific officer). There is no standard says West. “I’ve been using the Guardant 360 test [from Guardant Health, a US firm], but I’m just trying to get an early sense of how well it works,” he says. “My institution is interested in doing a trial comparing several companies that test serum with the tissue-based next generation sequencing we’re doing. There is no default company or test in the US, and I don’t think there will be, just as there isn’t one company doing next generation sequencing on tissue. But in the next few years we’ll likely see many larger institutions identify a test or company of choice for doing serum-based mutation testing.”
Ciardiello cautions on the technology, which will be in the hands of often hard-pressed pathology departments coming to terms with rapidly changing molecular pathology techniques, presenting challenges for both quality control and access. Initiatives that have assessed and accredited labs to carry out tissue-based molecular testing, such as for RAS in colorectal cancer (see Testing the testers, Cancer World Nov–Dec 2012) could be a model for serum testing. But many patients are still not accessing these tissue tests – indeed Ciardiello helped to launch the International Colorectal Cancer Association’s ‘Get Tested’ campaign this year, which aims to raise patients’ awareness of the importance of the test.
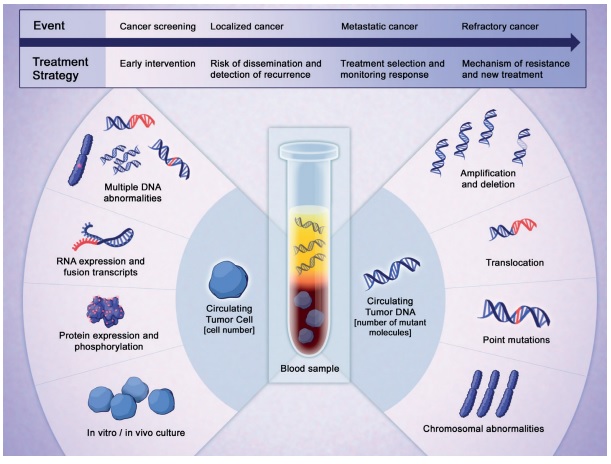
Liquid assets. Learning how to isolate and interpret the clues that solid tumours leave in the blood and other fluids could transform the way we detect cancers, select treatments and monitor response.
Reprinted with permission from AACR
A diagnostic tool
Rosenfeld agrees with Ciardiello that, while circulating DNA could be a useful prognostic tool indicating the presence of micrometastases after resection of the primary tumour, the greater immediate interest lies in its potential for revealing the evolving genetic profile of the tumour. “What’s moving most rapidly is performing cancer genomic analysis on a blood sample with a view to targeting mutations, because the research community wants to use genomics as a diagnostic tool,” he says. “Taking a tissue sample and testing it with low-sensitivity genomic analysis is analogous to carrying out high-sensitivity analysis on blood, and there will be particular patients and populations where either method could work better. You are asking the same question from the blood as you would from the tumour tissue.”
But if there are no actionable (i.e. targetable) mutations, or if treatment has changed mutations to be non-actionable, DNA can still be used as a highly sensitive monitoring tool, because even a few molecules are specific to the cancer, he adds. “This uses genomic techniques to obtain a quantitative measure, which then functions like a classic (e.g. protein) biomarker. The evidence so far supports the intuition that a higher DNA level is a bad sign, and that you can identify a recurrence earlier by seeing tumour DNA in the blood than by other means.”
He notes further work following the Johns Hopkins study, mostly more proof-of-concept studies on quantifying DNA levels and monitoring, including research at his own lab. While there is value in prognostics, he points out that it is more difficult to apply monitoring because there is a need to have pre-defined criteria for tumour progression, which would need randomised trials to establish.
Meanwhile, his lab and others are focusing more now on circulating DNA as a genomic research tool in cancer evolution and emerging mutations, resistance to therapy, and also earlier diagnosis, where adding genomic information from body fluid analysis to other symptoms could prove helpful.
A key paper from Rosenfeld’s lab was published in Nature in 2013 (vol 497, pp108–112) under the title ‘Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA’. This proof-of-principle study followed six patients, who had advanced breast, ovarian or lung cancers, for up to two years. The genomic evolution of these tumours included several types of mutations that were identified when treated with drugs such as paclitaxel, cisplatin, trastuzumab and gefitinib. The researchers concluded: “Serial analysis of cancer genomes in plasma constitutes a new paradigm for the study of clonal evolution in human cancers.” Other studies have since demonstrated similar results with more patients.
Impact on care
The impact is already being felt in clinical practice, at least in some research institutes. A good example comes from Gerhardt Attard’s group at the Institute of Cancer Research in London, where liquid biopsies are being used to inform the management of some patients. Research on men with advanced prostate cancer who receive the combination therapy of abiraterone (an enzyme inhibitor that decreases testosterone) and prednisone (a steroid that reduces inflammation), after having become resistant to initial hormone therapy, has shown that about one in five relapse on the combination regimen, because at some point androgen receptor mutations emerge that are activated by the steroid.
“This has a real, immediate impact on care, because we are now conducting trials where we stop prednisone as soon as we see the abnormality emerging, using a blood test do this. Importantly, prednisone is a very effective drug and works, at least initially, for most men, so it’s an example where the cancer adapts to turn an excellent drug into a driver of the disease in a small proportion of patients,” says Attard.
“We stop prednisone when we see the abnormality emerging, using a blood test to do this”
Beyond the immediate implications for patient care, the research done by Attard’s team has revealed important information about the behaviour of advanced prostate cancer and the complex ways in which abnormalities emerge, some of which is outlined in their paper (Sci Trans Med 2014, 6:254) on ‘Tumour clone dynamics in lethal prostate cancer’ (where ‘clone’ means a group of cells that share common changes).
This work involved looking at different abnormalities in the DNA. “We don’t restrict ourselves to mutations, but have also developed an approach to study changes in copy number, both amplifications and deletions,” he says. What his team found was that different groups of clones were circulating, suggesting lethal prostate cancer represents multiple different clones. “The variety and dynamic changes of DNA we detected in circulation were sobering,” says Attard. The team also found events that had previously been thought to be early, ‘initiating’, events in prostate cancer, analogous to mutations of the APC (adenomatous polyposis coli) gene in colorectal cancer, actually happened later.
Attard concludes that, critically, the ‘actionable’ aspect of such genomic work must be its link to treatment: “Clinical practice will be significantly changed for discoveries that show the utility of a test like this for predicting whether a patient will benefit or not from a treatment.”
West agrees, and says that newer drugs are likely to be the driver in his field, lung cancer, and that the barriers will come down in the next two to three years. “Currently there is not a pressing need to manage the standard of care with rebiopsy, but that will change with the introduction of the third-generation inhibitors, merelitinib and rociletinib, for EGFR mutation-positive patients with acquired resistance, which will require documentation of T790M positivity [T790M is a secondary mutation that limits the effectiveness of EGFR agents such as gefitinib].
“This will create a market and great need and value for serum-based testing. The other barrier is that there aren’t enough large-scale studies proving sensitivity and reliability of the assays, but these are being published now and in the coming year or two.”
There are many announcements about liquid biopsies now, and also collaborations starting up, such as Cancer-ID, a public–private consortium supported by Europe’s Innovative Medicines Initiative, which aims to validate blood-based cancer biomarkers for DNA, RNA and cells. Its academic leads are Klaus Pantel, based at the University Medical Centre Hamburg-Eppendorf, Germany, and Leon Terstappen, University of Twente, Netherlands (who developed CellSearch). Terstappen says that it will build on the CTC Trap project organised at his centre, which, as its name suggests, has been isolating and studying circulating tumour cells (see www.utwente.nl/tnw/ctctrap and www.cancer-id.eu).
In the US, the National Institutes of Health has recently reported work on the potential of circulating DNA to predict recurrence of the most common type of lymphoma. But it is the potential for early detection of pancreatic cancer that has really caught the attention of the media. Researchers have found a protein associated with circulating cancer exosomes (vesicles containing proteins, and DNA and other nucleic acids), and they were able to detect these exosomes, which carry specific KRAS mutations for pancreatic cancer, to “absolute specificity and sensitivity” in both late- and early-stage patients (Nature 2015, doi:10.1038/nature14581).
It is the potential for early detection of pancreatic cancer that has really caught the attention of the media
There were only five patients at early stage, however, and questions about whether there could be a screening test for early-stage pancreatic cancer remain unanswered, as there would be false-positives and negatives in the ‘real world’, which could mean unnecessary high-risk surgery, and pancreatic cancers still have a less than 20% five-year survival rate, even when detected at an early stage.
The research community is nonetheless excited about the potential of liquid biopsies for screening and early diagnosis, particularly in high risk groups. Some say this is where the greatest impact of liquid biopsies will be, despite the rapid progress in blood-based diagnostics to inform drug selection and disease management. Not least, there is potential for increasing our understanding of the way cancer evolves and metastases develop.





